Our research
We study epithelial cells, which acts as a physical barrier between the body and the environment. These cells are important in the defense against pathogens, and they are frequently bombarded with toxins, mutagens, and other noxious substances. It is of no surprise that most cancers arise from epithelial cells. The small intestine and colon are the most active and dynamic epithelial organs in he body. They are exposed to ~100 trillion microbes residing in the lumen of the gut on the apical side, and a well-developed immune system on the basal side in the underlying mucosa. The intestinal and colonic epithelia turn over on a weekly basis. Given the dynamic tasks of constant renewal, sensing of microbial and dietary changes, and responding to immune and stromal cell cues, it is amazing that the gut epithelium can maintain robust structure and function. We are interested in mechanisms of how epithelial cells handle and process different environmental cues to come up with coherent cell decisions to maintain homeostasis. We also also interested in how these processes go awry in chronic conditions such as Inflammatory Bowel Disease and colorectal neoplasms.
Cellular plasticity
Cellular plasticity refers to the ability of cell to transition from one unstable state to another. These transitions occur in committed cells that allow them to regain stem properties (dedifferentiation) or adopt a state that is completely non-native to the organ (cellular metaplasia). We investigate the mechanisms that trigger cellular plasticity, which can confer a neoplastic state and affect the downstream tumor microenvironment. We are particular interested in cell-microenvironment interactions that govern these transitions, and stemness in tumor cells that originate from normal stem and non-stem cells.
Microbiome-sensing cell types
We are interested in how specialized populations of microbe-sensing and responding epithelial cell types, such as tuft and Paneth cells, are specified and maintain their functions. We also investigate the dysfunction of these cell types under dysbiotic conditions and how the membership/function/localization of the microbiome is affected when these cells are perturbed.
In vivo systems biology
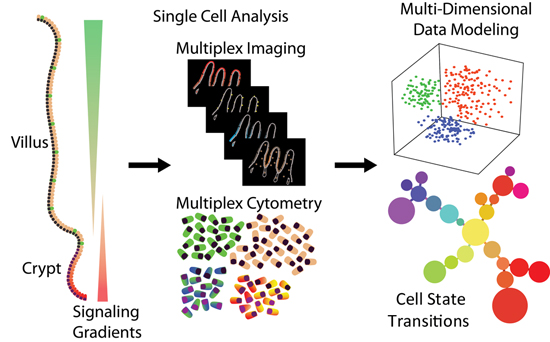
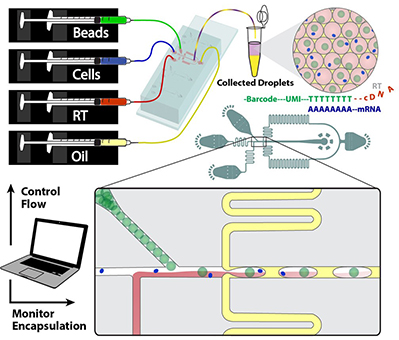
Integrative multi-modal analysis
We develop on various microfluidic platforms technologies to enable collection of high quality data via single-cell RNA-sequencing, spatial transcriptomics, multiplex microscopy imaging, and spatially resolved genomics. We integrate these orthogonal data collected on the same tissue specimens in order to maximize information content and paint comprehensive single-cell resolution picture of intercellular interactions.
Computational Biology
We develop data-science computational algorithms, when no others are available, to reveal biological insights that are latent in large datasets. Please visit our Github Repository.